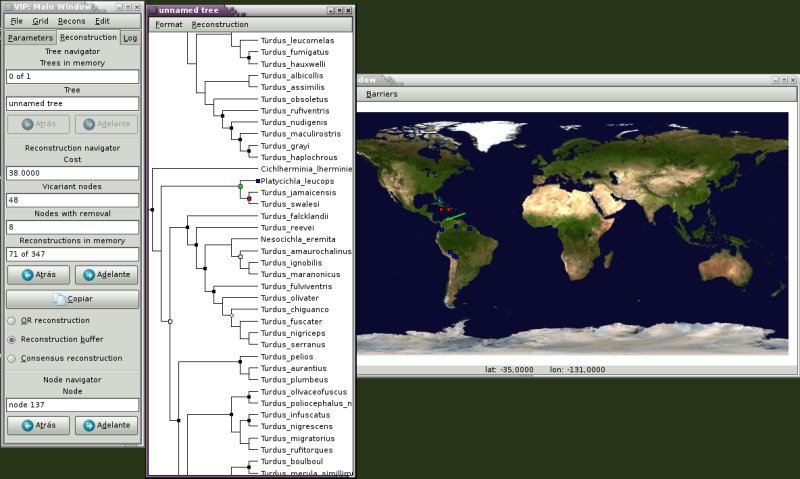
VIP is a free program for historical biogeography analysis. It uses geographical information in a phylogenetic context. The program is based on a Graphic User Interface that allows visualization and manipulation of biogeographic data.
VIP is developed for Spatial Vicariance Analysis [1]. This method was based on the procedure proposed by Peter Hovenkamp [2],[3]. The main goal of Spatial Vicariance Analysis is to detect disjoint distributions among sister groups. Disjoint (vicariant, allopatric) distributions provide information about barriers that separate populations, and eventually lead to speciation.
For several reasons, allopatry among actual taxa is not always straightforward. VIP takes into account partial overlapping among distributions, as well as a heuristic detection of overlapping based on removal of distribution on some nodes [4]. To make reconstructions comparable, an optimality criterion based on the number of vicariant nodes and the number of removed distributions is implemented in VIP.
For a full explanation on the method, see [1]. Also, the reading of Hovenkamp's papers [2],[3] and a classic paper of Don Rosen [5] is strongly recommended. You can download The VIP user's manual here, or a VIP primer as a quick introduction to the method (at the moment the primer is incomplete, but nevertheless I hope it will be useful).
Before downloading VIP, make sure that your system provides GTK support. Windows users can download the GTK runtime library here. If you have a problem with this GTK runtime, try this one (hat tip to Jonathan Liria)! Linux users, can download the GTK-bin library from their preferred repository, systems based on Gnome desktop system (such as Ubuntu or Debian) have GTK library already included.
If you are a TNT user, you can save your tree to be read with VIP using this macro. The parameter asking for the script is the tree to be saved; if no tree is provided, it uses the tree 0. The results are saved as tree.xml.
Geospatial data can be read from KML (Google earth) format, Darwin core format (GBIF standard), or Tab delimited text.
Raster images in jpeg, tif, png, bmp and gif format can be read.

VIP running under linux. Map from NASA blue marble.
Please cite VIP when using the program in your own work. The suggested citation is:
Arias, J. S. 2010. VIP: Vicariance Inference Program, Software and documentation published by the author. Available at: http://www.zmuc.dk/public/phylogeny/vip.
Also, do not forget to cite the main method description [1], and Hovenkamp's papers [2],[3]. Several features are based on other people's research, and they must be recognized if you use them: [4],[6], [7].
[1] Arias, J.S., Szumik, C.A., Goloboff, P.A. In press. Spatial analysis of vicariance: A method for using direct geographical information in historical biogeography. Cladistics. DOI: 10.1111/j.1096-0031.2011.00353.x.
[2] Hovenkamp, P. 1997. Vicariance events, not areas, should be used in biogeographical analysis. Cladistics 13: 67. DOI: 10.1111/j.1096-0031.1997.tb00241.x.
[3] Hovenkamp, P. 2001. A direct method for the analysis of vicariance patterns. Cladistics 17: 260. DOI: 10.1006/clad.2001.0176.
[4] Page, R.D.M. 1994. Parallel phylogenies: Reconstructing the history of host-parasite assemblages. Cladistics 10: 155. DOI: 10.1111/j.1096-0031.1994.tb00170.x.
[5] Rosen, D.E. 1978. Vicariant patterns and historical explanation in biogeography. Systematic Zoology 27: 159. DOI:10.2307/2412970.
[6] Hunn, C.A., Upchurch, P. 1978. The importance of time/space in diagnosing the causality of phylogenetic events: Towards a 'chronobiogeographical' paradigm? Systematic Biology 50: 391. DOI:10.1093/sysbio/50.3.391.
[7] Bremer, K. 1988. The limits of amino-acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42: 795-803. JSTOR: 2498870.